
Computer technology helps to make timber identification easier
Naturalis Biodiversity CenterForests cover 30 percent of the earth's land area, representing about four billion hectares and three trillion trees. Over the past fifteen years, forested regions comparable to the combined area of France, Spain and the United Kingdom have been lost worldwide, which has given rise to great concern. This loss both reduces the carbon storage of forests – one of our main buffers of CO2 extracted from the atmosphere – and leads to alarming declines in biodiversity. Recent evaluations of global logging show that deforestation in the tropics is currently occurring at an even more worrying pace, due to unsustainable agriculture, mining and illegal felling. However, illegal logging rarely leads to prosecution, as few efficient forensic tools are available for timber identification.
Wood anatomy remains the most commonly used method for the taxonomic identification of tree trunks. It takes years of training to become an expert in this field, however, leading to fewer wood anatomists with extensive experience. In addition, wood anatomists can generally identify wood at the level of the genus rather than the species, while the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) often requires species-level identification. For this reason, wood anatomists and computer scientists are now joining forces to develop faster and more accurate methods of identifying wood samples through computer-aided support for microscopic wood photographs.
Classification at the species level
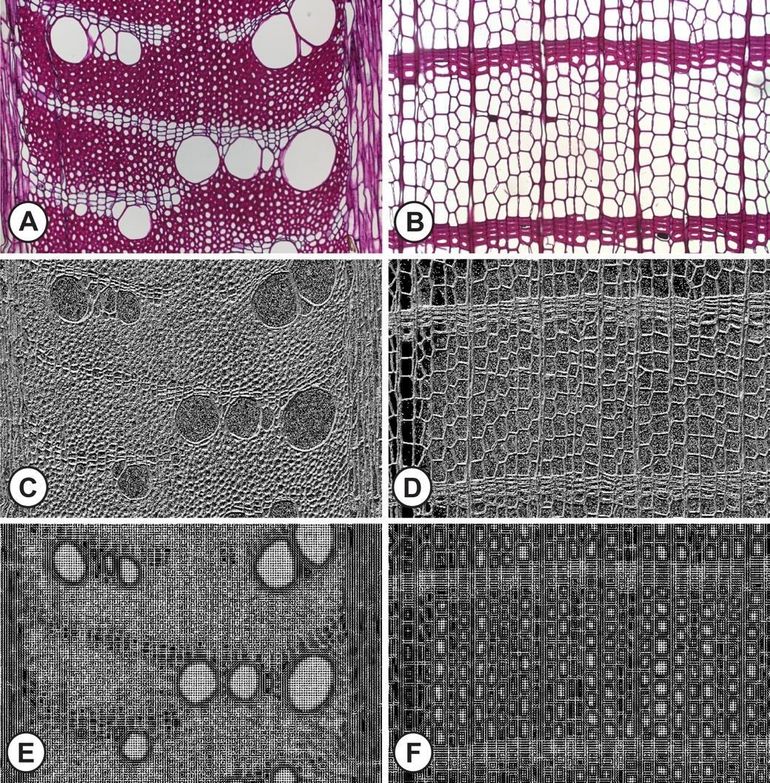
Based on an existing database of microscopic cross-sectional images (figures A & B below), with 20 microscopic images for each of the 112 wood species examined, the researchers classified the images at the species level on the basis of computer-generated features (figures C-F below) that were compared to more advanced methods based on Deep Learning or Convolutional Neural Networks (CNNs). Thanks to the Deep Learning approach, the recognition percentage of the species has increased to as much as 96.4 percent. This offers opportunities to expand the available photo dataset with new cross-sectional and longitudinal images of all tree species on the CITES list, as well as the species that demonstrate strong wood-anatomical similarities with the CITES species. Thanks to this new reference database, it will be possible in the future to develop a tool that enables customs officials and other stakeholders to identify a block of wood even more accurately than experienced wood anatomy experts.
More information
- Read the whole aricle for more information: Computer-assisted timber identification based on features extracted from microscopic wood sections.
Text: Frederic Lens, Mehrdad Jahanbanifard en Barbara Gravendeel, Naturalis Biodiversity Center & Fons Verbeek, Leiden Institute of Advanced Computer Science, LIACS, Universiteit Leiden
Photos: Getty Images; Naturalis
